A high-performance Rust implementation of the Turnbull non-parametric maximum likelihood estimator for interval-censored survival data
Warning! I am not a statistician. This article is not reviewed. Please confer with a responsible adult!
A common need in biostatistics is to estimate survival curves, but particular difficulty arises when observations are interval censored, i.e. the time of event is not observed exactly, but is known only to fall within a particular interval. In this setting, the Turnbull estimator [1] extends the usual Kaplan–Meier estimator to interval-censored data.
There are several publicly available software implementations for fitting the Turnbull estimator. These include icenReg and interval (using Icens and MLEcens) for R, and lifelines and surpyval for Python. Software for fitting interval-censored Cox regression can also be trivially applied to fit the Turnbull estimator, such as stintcox for Stata 17+. Of these, only interval supports computing standard errors, which it achieves via a computationally expensive bootstrap.
hpstat turnbull is a Rust implementation of the Turnbull estimator using an efficient expectation maximisation–iterative convex minorant algorithm described by Anderson-Bergman [2], who also implements this in icenReg. Standard errors are estimated in a computationally efficient manner using the Wald method or by inverting likelihood ratio tests [3]. This contrasts with the bootstrap-based approach used by interval.
Performance
To evaluate the performance of hpstat turnbull compared with other software implementing the Turnbull estimator, we fit a model on a real-world dataset comprising 16639 observations and 1546 time points. Software was run using the default options, except Stata and interval, whose stintcox and icfit respectively did not converge within 5 minutes using default options, nor did icfit using initfit='initEMICM' from Icens. The table below shows the model log-likelihoods obtained by the software.
| Software | Version | Options | Model log-likelihood |
|---|---|---|---|
| hpstat | 8914cf3 | --se-method oim-drop-zeros --output json | −20916.31992 |
| icenReg | 2.0.15 | −20916.31 | |
| Stata | 17.0 | favorspeed | −21003.834 |
| lifelines | 0.27.7 | −20916.36 | |
| interval | 1.1-1.0 | initfit='initcomputeMLE' (from MLEcens) | −20916.31 |
| surpyval | 0.10.10 | −20918.41 |
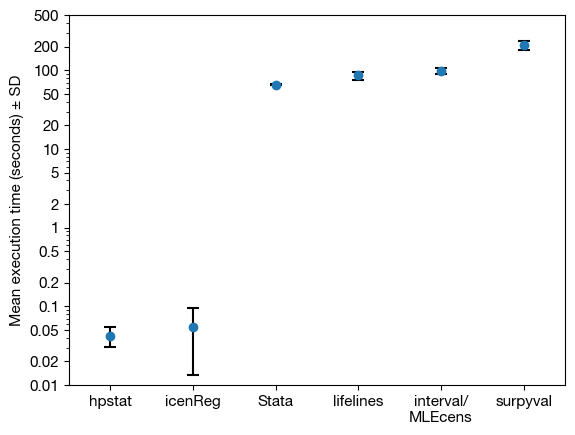
The figure below shows the mean execution times on an Intel Core i5-7500. Error bars are shown at 10× true scale. Standard errors for survival probabilities were computed only in hpstat.
Discussion
hpstat turnbull and icenReg were the fastest-performing implementations, with hpstat slightly faster and additionally computing standard errors. Both were significantly faster than all other software in this comparison. It is noted, however, that hpstat has not been extensively reviewed or tested.
hpstat is available at https://yingtongli.me/git/hpstat.
References
- Turnbull BW. The empirical distribution function with arbitrarily grouped, censored and truncated data. Journal of the Royal Statistical Society, Series B (Methodological). 1976;38(3):290–5. doi: 10.1111/j.2517-6161.1976.tb01597.x
- Anderson-Bergman C. An efficient implementation of the EMICM algorithm for the interval censored NPMLE. Journal of Computational and Graphical Statistics. 2017;26(2):463–7. doi: 10.1080/10618600.2016.1208616
- Goodall RL, Dunn DT, Babiker AG. Interval-censored survival time data: confidence intervals for the non-parametric survivor function. Statistics in Medicine. 2004;23(7):1131–45. doi: 10.1002/sim.1682